This page was generated by
nbsphinx from
docs/notebooks/analysis/swept_langmuir/find_floating_potential.ipynb.
Interactive online version:
.
Swept Langmuir Analysis: Floating Potential
This notebook covers the use of the find_floating_potential() function and how it is used to determine the floating potential from a swept Langmuir trace.
The floating potential, \(V_f\), is defined as the probe bias voltage at which there is no net collected current, \(I=0\). This occurs because the floating potential slows the collected electrons and accelerates the collected ions to a point where the electron- and ion-currents balance each other out.
[1]:
%matplotlib inline
from pathlib import Path
import matplotlib.pyplot as plt
import numpy as np
from plasmapy.analysis import swept_langmuir as sla
plt.rcParams["figure.figsize"] = [10.5, 0.56 * 10.5]
np.set_printoptions(precision=4, threshold=16)
Contents:
How find_floating_potential() works
The passed current array is scanned for points that equal zero and point-pairs that straddle where the current, \(I\), equals zero. This forms an collection of “crossing-points.”
The crossing-points are then grouped into “crossing-islands” based on the
thresholdkeyword.A new island is formed when a successive crossing-point is more (index) steps away from the previous crossing-point than defined by
threshold. For example, ifthreshold=4then a new island is formed if a crossing-point candidate is more than 4 steps away from the previous candidate.If multiple crossing-islands are identified, then the function will compare the total span of all crossing-islands to
min_points. If the span is greater thanmin_points, then the function is incapable of identifying \(V_f\) and will returnnumpy.nanvalues; otherwise, the span will form one larger crossing-island.
To calculate the floating potential…
If the number of points that make up the crossing-island is less than
min_points, then each side of the “crossing-island” is equally padded with the nearest neighbor points untilmin_pointsis satisfied.If
fit_type="linear", then ascipy.stats.linregressfit is applied to the points that make up the crossing-island.If
fit_type="exponential", then ascipy.optimize.curve_fitfit is applied to the points that make up the crossing-island.
Notes about usage
The function provides no signal processing. If needed, the user must smooth, sort, crop, or process the arrays before passing them to the function.
The function requires the voltage array to be monotonically increasing.
If the total range spanned by all crossing-islands is less than or equal to
min_points, thenthresholdis ignored and all crossing-islands are grouped into one island.
Knobs to turn
fit_typeThere are two types of curves that can be fitted to the identified crossing point data:
"linear"and"exponential". The former will fit a line to the data, whereas, the latter will fit an exponential curve with an offset. The default curve is"exponential"since swept Langmuir data is not typically linear as it passes through \(I=0\).min_pointsThis variable specifies the minimum number of points that will be used in the curve fitting. As mentioned above, the crossing-islands are identified and then padded until
min_pointsis satisfied. Usage:min_points = None(DEFAULT):min_pointsis chosen to be the larger of 5 orfactor * array_size, wherefactor = 0.1for"linear"and0.2for"exponential".min_points = numpy.inf: The entire array is fitted.min_pointsis an integer>= 1:min_pointsis the minimum number of points to be used.0 < min_points < 1: The minimum number of points is taken asmin_points * array_size.
thresholdThe max allowed index distance between crossing-points before a new crossing-island is formed.
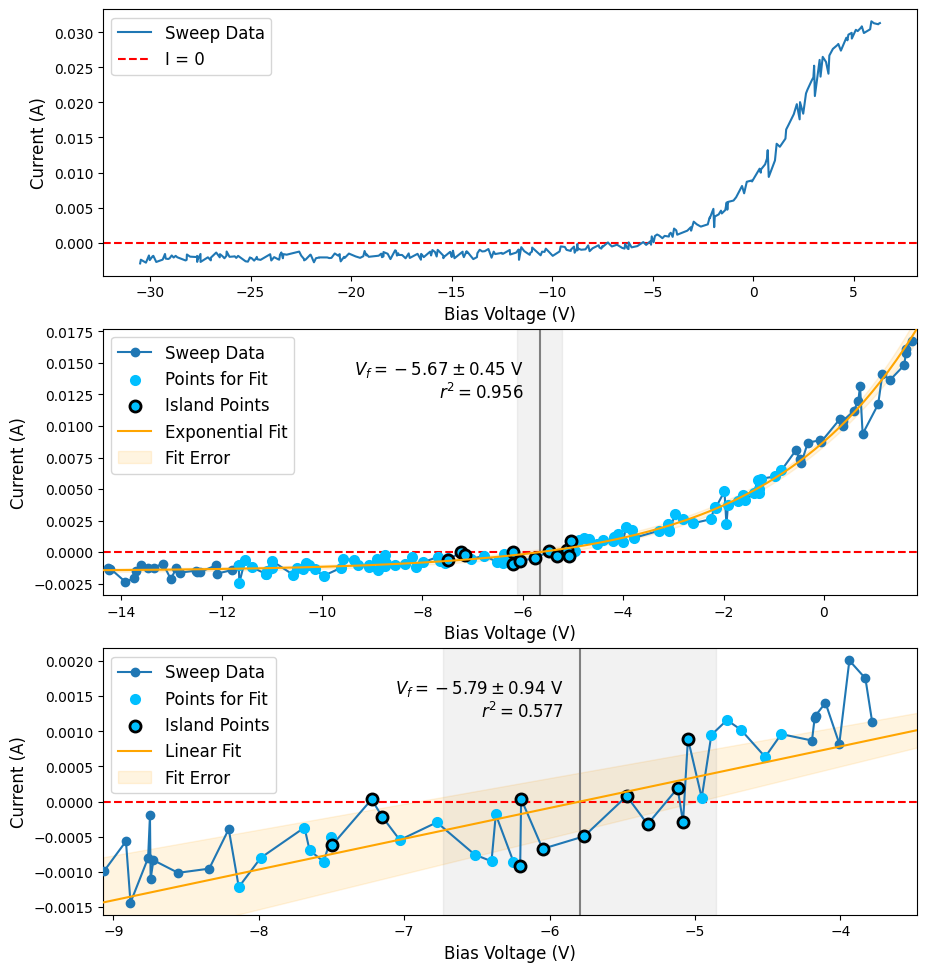
Calculate the Floating Potential
Below we’ll compute the floating potential using the default fitting behavior (fit_type="exponential") and a linear fit (fit_type="linear").
[2]:
# load data
filename = "Beckers2017_noisy.npy"
filepath = (Path.cwd() / ".." / ".." / "langmuir_samples" / filename).resolve()
voltage, current = np.load(filepath)
# voltage array needs to be monotonically increasing/decreasing
isort = np.argsort(voltage)
voltage = voltage[isort]
current = current[isort]
# get default fit results (exponential fit)
results = sla.find_floating_potential(voltage, current, min_points=0.3)
# get linear fit results
results_lin = sla.find_floating_potential(voltage, current, fit_type="linear")
Interpreting results
The find_floating_potential() function returns a 2 element tuple, where the first element is the calculated floating potential \(V_f\) and the second element is a named tuple VFExtras containing additional parameters resulting from the calculation.
results[0]is the determined floating potential (same units as the passvoltagearray)
[3]:
vf = results[0]
vf
[3]:
-5.665730698520963
results[1]is an instance of VFExtras and contains additional information from the calculation
[4]:
extras = results[1]
extras
[4]:
VFExtras(vf_err=0.45174119664601947, rsq=0.9564766142727993, fitted_func=f(x) = a exp(alpha x) + b <class 'plasmapy.analysis.fit_functions.ExponentialPlusOffset'>, islands=[slice(192, 195, None), slice(201, 210, None)], fitted_indices=slice(155, 247, None))
extras[0]=extras.vf_err= the associated uncertainty in the \(V_f\) calculation (same units asvf)
[5]:
(extras[0], extras.vf_err)
[5]:
(0.45174119664601947, 0.45174119664601947)
extras[1]=extras.rsq= the coefficient of determination (r-squared) value of the fit
[6]:
(extras[1], extras.rsq)
[6]:
(0.9564766142727993, 0.9564766142727993)
extras[2]=extras.fitted_func= the resulting fitted functionextras.fitted_funcis a callable representation of the fitted functionI = extras.fitted_func(V).extras.fitted_funcis an instance of a sub-class ofAbstractFitFunction. (FitFunction classes)Since
extras.fitted_funcis a class instance, there are many other attributes available. For example,extras.fitted_func.paramsis a named tuple of the fitted parametersextras.fitted_func.param_errorsis a named tuple of the fitted parameter errorsextras.fitted_func.root_solve()finds the roots of the fitted function. This is how \(V_f\) is calculated.
[7]:
(
extras[2],
extras.fitted_func,
extras.fitted_func.params,
extras.fitted_func.params.a,
extras.fitted_func.param_errors,
extras.fitted_func.param_errors.a,
extras.fitted_func(vf),
)
[7]:
(f(x) = a exp(alpha x) + b <class 'plasmapy.analysis.fit_functions.ExponentialPlusOffset'>,
f(x) = a exp(alpha x) + b <class 'plasmapy.analysis.fit_functions.ExponentialPlusOffset'>,
FitParamTuple(a=0.010275375458525872, alpha=0.338277953104458, b=-0.0015115846190350466),
0.010275375458525872,
FitParamTuple(a=0.00031439926674768734, alpha=0.02163110937712501, b=0.00012999596409070833),
0.00031439926674768734,
0.0)
extras[3]=extras.islands= a list of slice objects representing all the identified crossing-islands
[8]:
(
extras[3],
extras.islands,
voltage[extras.islands[0]],
)
[8]:
([slice(192, 195, None), slice(201, 210, None)],
[slice(192, 195, None), slice(201, 210, None)],
array([-7.496 , -7.2238, -7.1512]))
extras[4]=extras.fitted_indices= a slice object representing the indices used in the fit
[9]:
(
extras[4],
extras.fitted_indices,
voltage[extras.fitted_indices],
)
[9]:
(slice(155, 247, None),
slice(155, 247, None),
array([-11.6594, -11.6488, -11.5358, ..., -1.251 , -0.9812, -0.8504]))
Plotting results
[10]:
figwidth, figheight = plt.rcParams["figure.figsize"]
figheight = 2.0 * figheight
fig, axs = plt.subplots(3, 1, figsize=[figwidth, figheight])
# plot original data
axs[0].set_xlabel("Bias Voltage (V)", fontsize=12)
axs[0].set_ylabel("Current (A)", fontsize=12)
axs[0].plot(voltage, current, zorder=10, label="Sweep Data")
axs[0].axhline(0.0, color="r", linestyle="--", label="I = 0")
axs[0].legend(fontsize=12)
# zoom on fit
for ii, label, rtn in zip([1, 2], ["Exponential", "Linear"], [results, results_lin], strict=False):
vf = rtn[0]
extras = rtn[1]
# calc island points
isl_pts = np.array([], dtype=np.int64)
for isl in extras.islands:
isl_pts = np.concatenate((isl_pts, np.r_[isl]))
# calc xrange for plot
xlim = [voltage[extras.fitted_indices].min(), voltage[extras.fitted_indices].max()]
vpad = 0.25 * (xlim[1] - xlim[0])
xlim = [xlim[0] - vpad, xlim[1] + vpad]
# calc data points for fit curve
mask1 = np.where(voltage >= xlim[0], True, False)
mask2 = np.where(voltage <= xlim[1], True, False)
mask = np.logical_and(mask1, mask2)
vfit = np.linspace(xlim[0], xlim[1], 201, endpoint=True)
ifit, ifit_err = extras.fitted_func(vfit, reterr=True)
axs[ii].set_xlabel("Bias Voltage (V)", fontsize=12)
axs[ii].set_ylabel("Current (A)", fontsize=12)
axs[ii].set_xlim(xlim)
axs[ii].plot(
voltage[mask],
current[mask],
marker="o",
zorder=10,
label="Sweep Data",
)
axs[ii].scatter(
voltage[extras.fitted_indices],
current[extras.fitted_indices],
linewidth=2,
s=6**2,
facecolors="deepskyblue",
edgecolors="deepskyblue",
zorder=11,
label="Points for Fit",
)
axs[ii].scatter(
voltage[isl_pts],
current[isl_pts],
linewidth=2,
s=8**2,
facecolors="deepskyblue",
edgecolors="black",
zorder=12,
label="Island Points",
)
axs[ii].autoscale(False)
axs[ii].plot(vfit, ifit, color="orange", zorder=13, label=label + " Fit")
axs[ii].fill_between(
vfit,
ifit + ifit_err,
ifit - ifit_err,
color="orange",
alpha=0.12,
zorder=0,
label="Fit Error",
)
axs[ii].axhline(0.0, color="r", linestyle="--")
axs[ii].fill_between(
[vf - extras.vf_err, vf + extras.vf_err],
axs[1].get_ylim()[0],
axs[1].get_ylim()[1],
color="grey",
alpha=0.1,
)
axs[ii].axvline(vf, color="grey")
axs[ii].legend(fontsize=12)
# add text
rsq = extras.rsq
txt = f"$V_f = {vf:.2f} \\pm {extras.vf_err:.2f}$ V\n"
txt += f"$r^2 = {rsq:.3f}$"
txt_loc = [vf, axs[ii].get_ylim()[1]]
txt_loc = axs[ii].transData.transform(txt_loc)
txt_loc = axs[ii].transAxes.inverted().transform(txt_loc)
txt_loc[0] -= 0.02
txt_loc[1] -= 0.26
axs[ii].text(
txt_loc[0],
txt_loc[1],
txt,
fontsize="large",
transform=axs[ii].transAxes,
ha="right",
)